KRISS Identifies a Major Nuclease Required for the Degradation of Damage-containing DNA Fragments
- Writerkrissadmin
- Date2022-05-11 00:00
- Hits743
KRISS Identifies a Major Nuclease Required for the Degradation of Damage-containing DNA Fragments
- KRISS finds the cause of the decrease in damage-containing DNA fragments produced in response to UV and carcinogens -
- Highly sensitive technique for measuring DNA fragments will help research on personalized cancer treatments -
The Korea Research Institute of Standards and Science (KRISS, President Hyun-Min Park) found that the TREX1 protein identified in a genetic screen degrades damage-containing DNA fragments, successfully proving this in vitro.

▲ KRISS researchers who identified the TREX1 enzyme as a major regulator of damage-containing DNA oligonucleotides in vivo (from left: Yunju Song, student researcher; Jun-Hyuk Choi, principal researcher; Geun Hoe Kim, UST student researcher)
DNA damage occurs every day due to UV radiation and due to chemical carcinogens as well. Genetic information is preserved in cells despite such damage because cells have a repair mechanism for damaged bases. DNA damage accumulates if DNA repair is not effective, leading to aging or serious diseases such as cancer.
Damage-containing DNA oligonucleotides are inevitably produced from the DNA repair process. These DNA fragments decrease gradually over time in human cells. If they are not properly regulated, it can cause diseases through inflammation or inappropriate immune reactions. In earlier research, there was no technology capable of analyzing small traces of DNA fragments, and the causes of related decreases remained unknown. The KRISS research team found that the TREX1 protein contributes to the degradation of damage-containing DNA fragments, becoming the world’s first team to prove this in vitro.
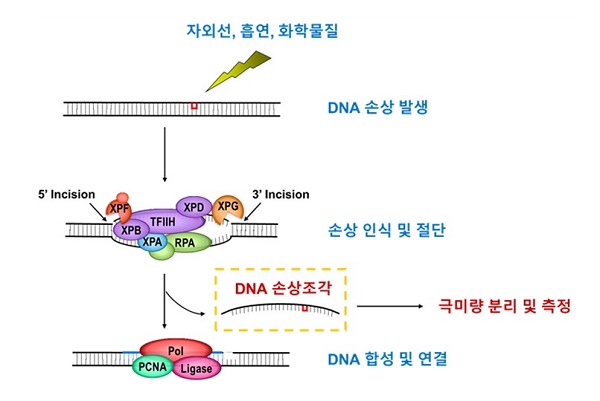
▲ Measuring damage-containing DNA fragments produced during the DNA repair process
The research team altered the expression of specific target genes in human cells and then analyzed the abundance of damage-containing DNA fragments. They showed that increasing amounts of TREX1 significantly reduced the amounts of damage-containing DNA fragments. They also purified large amounts of damage-containing DNA along with recombinant TREX1 and showed that TREX1 degrades the damage-containing DNA in vitro.
Jun-Hyuk Choi, principal researcher of the KRISS Biometrology Group, said, “DNA fragments will cause aging and diseases if not properly regulated. The persistent presence of DNA fragments in cancer cells is associated with the development of resistance to cancer therapeutics. This study of the degradation of DNA fragments will be very useful for anti-cancer researchers.”
The team’s success was made possible by KRISS’ highly sensitive techniques for measuring damage-containing DNA fragments in small numbers of cells. In 2015, KRISS developed a novel in vivo assay capable of detecting DNA fragments containing a wide spectrum of causes of DNA lesions (UV damage, chemical carcinogens, and chemotherapeutic drugs). The measurement technique has since been improved to allow high-precision analysis within three minutes of DNA damage. The sample amounts required for detection were also reduced to one-tenth of the original level, which is equivalent to roughly 10 picograms.
* Picogram: A unit of mass equal to 10-12 grams. An E.coli bacterium has a mass of approximately 1 picogram, and the DNA in the cells of humans in general weighs 6 picograms.

▲ KRISS researchers examined DNA damage after exposing cells to various carcinogens.
The unique technique allows for direct comparisons of individual DNA repair activities and thus facilitates personalized cancer treatments by accessing cancer risks or anticancer drug efficacy rates. KRISS plans to develop the technique further for measuring trace amounts of DNA repair products. Thus, the technology for measuring DNA repair activity in individuals will lay the foundation for future clinical applications.
Funded by KRISS and the National Research Foundation of Korea, the research outcomes were published in the prestigious journal Nucleic Acids Research (IF 16.97, first/corresponding author) on April 22. The paper was selected as a Hanbitsa (People Glorifying Korea) paper by the Biological Research Information Center (BRIC). The primary authors are Seon Hee Kim and Geun Hoe Kim, UST master’s students advised by Dr. Jun-Hyuk Choi, and the co-corresponding author is Professor Michael G. Kemp of Wright State University.

